Description
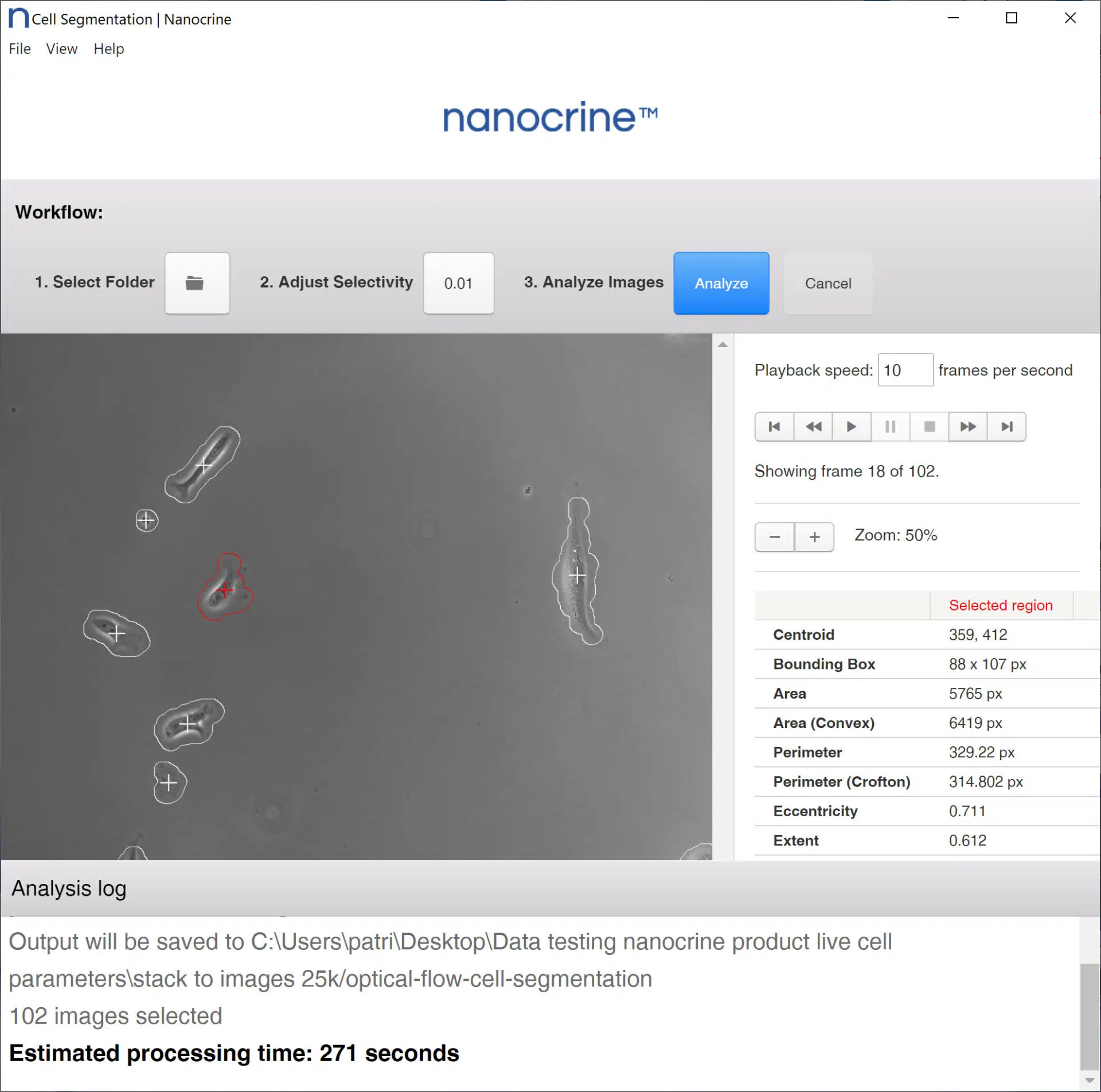
Nanocrine Cell Image Analyzer automates the process of cell image segmentation and data collection. Conventional methods for segmenting microscopy imagery usually involve manually identifying cells and marking their boundaries by hand. Cell Image Analyzer is the first software-based solution for cell data collection that uses cell motion to easily distinguish which portions of an image belong to a cell and which ones contain only background noise.
First, timelapse imagery is loaded into Cell Image Analyzer. Using an optical flow algorithm, Cell Image Analyzer then calculates the displacement of each object in view between one frame and the next. It ranks every pixel by the amount of displacement and filters out the areas of the image with low displacement. The final result is a black-and-white Cell Map for each image, where the white areas are cells and the black areas are background.
Cell Image Analyzer computes a wide variety of useful geometric cell parameters from the generated Cell Maps and outputs it as a CSV file, which, by itself, may be sufficient for a whole publication. However, the user is also free to import their Cell Maps into their computational software of choice, such as ImageJ.
For a limited time, Cell Image Analyzer is being offered as a free bonus for registering an account on Nanocrine.com. Sign up today and you will receive a one-time download link.
Key Features:
- Increased repeatability and accuracy in live-cell data
- Drastic reduction in time spent between data collection and publication
- Customizable displacement threshold so the user can fine-tune Cell Image Analyzer for their experimental setup
- Live preview of displacement threshold so the user can make adjustments and see a sample Cell Map before committing to processing their whole image set